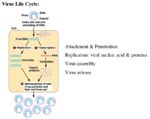
Viral Replication
- Virion attaches to host cell
- Virus penetrates into the cytoplasm
- Genome is uncoated
- Virus interacts with synthetic apparatus of the cell, producing viral proteins
- These proteins assemble into new virus particles, which then exit the cell
Viruses can only replicate in host cells as they do not possess the structures or the enzymes to replicate independently. When a virus utilises the host cell for replication, the host cell can experience effects ranging from minor changes in metabolism to lysis. In most cases, a virus replication life cycle takes between six to forty hours. During initial infection the host cell undergoes a period called the "eclipse" period where it is very difficult to detect infected cells. After this period the presence of a virus can usually be demonstrated both intracellularly and extracellularly. Post-eclipse period the number of viral particles present increases exponentially.
Virus replication can be divided into various stages;
1. Attachment and entry into the host cell
2. Uncoating of viral nucleic acid
3. Synthesis of virus-specific proteins
4. Production of new viral nucleic acid
5. Assembly and release of newly-formed viruses from the host cell
1. Attachment and Entry
Viruses must adhere to the host cell surface receptors in order to be infective. It is currently thought that the initial virus-host cell interaction is a purely random event related to the number of viral particles and susceptible host cells i.e. which cell membrane receptors they exhibit. Many species of virus are highly evolved and are able to attach to multiple types of receptors found on the surface of potential host cells. Uptake of the virus by the host cell is an energy dependant process and can occur in a number of ways. Receptor-mediated endocytosis may occur after attachment and the virus is taken into the cell in a vesicle. The envelopes surrounding some species such as orthomyxoviuses, rhabdoviuses and flaviviruses are able to interact with the membrane of the vesicle, fusing with it and then allowing the nuclear material to be released directly into the cytoplasm of the host cell. Another entry method used by species such as paramyxoviruses, retroviruses and herpesviruses involves the fusion of the envelope with the plasma membrane facilitating the release of the nuclear material into the host cell cytoplasm. Non-enveloped viruses such as picornaviruses use channels in the plasma membrane to directly enter the cytoplasm of the host cell.
2. Uncoating
This is the process whereby the viral nuclear material is released into the cell in a form that is suitable for transcription using the host cells structures and enzymes. Enveloped viruses that have discharged their nucleocapsid material into the cytoplasm of the cell are able to undergo transcription. Non-enveloped viruses have a less well understood mechanism for uncoating their capsid although it is thought that it involves lysosomal proteolytic enzyme activity. Retroviruses are able to undertake all of the same functions as other viruses but they undergo transcription without complete release from the capsid. Some viruses such as Poxviruses are uncoated in two stages with the initial stage being undertaken by host cell enzymes and the second stage requiring virus-specific proteins. In viruses that require their nuclear material to enter the host cell nucleus, the process of uncoating can be completed adjacent to a nuclear pore.
3. Synthesis
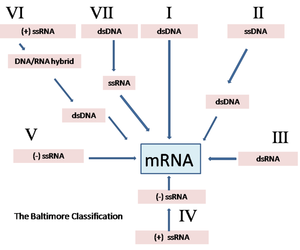
The viral genetic material is replicated using host structures and in most cases host enzymes produce mRNA. DNA viruses that undertake transcription within the nucleus utilise the host cell's transcriptases to facilitate replication whilst other viruses are able to utilise their own enzymes and the host cell's structures to produce viral mRNA. Due to the various ways in which viruses are able to undergo transcription to produce viral mRNA, viruses of veterinary importance have been grouped into six classes called the Baltimore Classification. The central theme of this classification is whether the virus is the designation of single-stranded RNA viruses into either positive-sense or negative-sense nucleic acid. The term "sense" refers to the polarity of the nucleic acid. The nucleic acid of positive-sense single-stranded RNA viruses is mRNA and this can be translated directly into viral protein rather than requiring to go through the process of transcription.
4. Replication of Viral Nucleic Acid
DNA Viruses
Viruses such as herpesvirus, papovavirus and adenovirus are double-stranded DNA viruses and replicate in the host cell nucleus. In this case the viral DNA is transcribed by the host cell RNA polymerase (transcriptase) forming mRNA. In contrast to this, single-stranded DNA viruses such as parvoviruses and circoviruses utilise host cell DNA polymerase whilst replicating in the host cell nucleus. This process results in the production of double-stranded DNA which is then transcribed to mRNA. In both cases the first genes replicated code for enzymes and proteins necessary for further stages of replication and the suppression of host cell genes that may prevent viral replication. Later produced proteins from the viral DNA often encode the actual viral nucleic acid. This sequence of genetic events is not proven to occur in RNA viruses.
RNA Viruses
Double-stranded RNA viruses such as Reoviruses and Birnaviruses undergo transcription in the cytoplasm of the host cell via viral transcriptase. Negative-sense strands (where they exist) are transcribed to produce individual mRNA molecules. Positive-sense, single-stranded RNA viruses are able to act directly as mRNA. The nucleic acid from RNA viruses is able to directly bind ribosomes and therefore in RNA viruses the naked genetic material itself is infective, rather than just the outer coatings of DNA viruses. It should be noted that differences between positive and negative-sense and single and double-stranded replication are far more complex than described above but this level of detail is beyond the scope of this overview.
5. Assembly and Release of Viruses
As expected given the differences between viral replication, there are a wide range of methods used by viruses for assembly and release of viruses. There are also differences caused by whether the virus is enveloped or non-enveloped. The structural proteins of non-enveloped viruses associate first forming a procapsid and the nucleic acid is then added to the procapsid. Non-enveloped viruses such as picornaviruses and reoviruses form in the cytoplasm of the host cell whereas other non-enveloped viruses such as parvoviruses, adenoviruses and papoviruses are assembled within the nucleus.
Enveloped viruses require a further stage of formation in comparison to non-enveloped viruses in that they must acquire the envelope. This is mainly done during the budding phase from the cell membrane. Prior to the budding phase, enveloped viruses modify the host cell membrane by inserting virus-specific transmembrane glycoproteins which aggregate in patches in the plasma membrane facilitating the budding process. However this process also has a side affect of altering the antigenic properties of the cell membrane making them a target for cytotoxic T lymphocytes. When budding occurs the plasma membrane is not breached and the resulting envelope surrounding the virus contains non-cytopathic properties which are difficult to detect by the host immune system. Therefore enveloped viruses are associated with persistent infections in more cases than non-enveloped viruses. Viruses such as flaviviruses, coronaviruses, arteriviruses and bunyaviruses acquire their envelopes inside the cells within the rough endoplasmic reticulum or the golgi apparatus. The viruses are then transported to the host cell membrane in vesicles which fuse with the membrane and are released by exocytosis.