Difference between revisions of "Major Histocompatability Complexes"
Jump to navigation
Jump to search
| (60 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | {{ | + | {{toplink |
| + | |backcolour = FFE4E1 | ||
| + | |linkpage =Immunology | ||
| + | |linktext =Immunology | ||
| + | |tablelink = Adaptive Immune System (Table) - WikiBlood | ||
| + | |sublink1 =Adaptive Immune System - WikiBlood | ||
| + | |subtext1 =ADAPTIVE IMMUNE SYSTEM | ||
| + | |sublink2=Lymphoreticular System - Anatomy & Physiology#Lymphocytes | ||
| + | |subtext2=LYMPHORETICULAR SYSTEM | ||
| + | |pagetype =Blood | ||
| + | }} | ||
| + | <br> | ||
[[Image:MHC.jpg|thumb|right|250px|Major Histocompatibility Complexes - B. Catchpole, RVC 2008]] | [[Image:MHC.jpg|thumb|right|250px|Major Histocompatibility Complexes - B. Catchpole, RVC 2008]] | ||
| − | + | =Introduction= | |
| − | T-cells rely on Major Histocompatability Complexes (MHC) | + | T-cells rely on Major Histocompatability Complexes (MHC) to present antigen fragments for their recognition. MHC has evolved to form two classes for antigen presentation: '''MHC I''' presents digestion fragments from antigen in '''cellular cytoplasm''', and '''MHC II''' presents digestion fragments from antigen in the '''tissue fluid'''. As such, MHC I tends to bind slightly smaller peptides (~9 amino acids) than MHC II (~15 amino acids). |
| + | |||
| + | =Classes= | ||
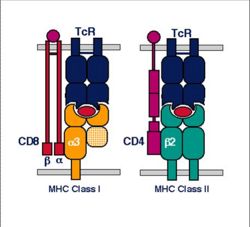
==MHC I== | ==MHC I== | ||
| + | [[Image:MHC I processing.jpg|thumb|200px|right|'''MHC I presentation pathway, courtesy of B. Catchpole, 2008''']] | ||
===Structure=== | ===Structure=== | ||
| − | + | * MHC class I is expressed on virtually all nucleated cells | |
| − | + | * MHC class I consists of a membrane-associated heavy chain bound non-covalently with a secreted light chain | |
| − | MHC class I is expressed on virtually all nucleated cells | + | ** Heavy chain: |
| − | + | *** Made up of three distinct extracellular protein domains | |
| − | + | **** α1, α2 and α3 | |
| − | The MHC class I domains are structurally and genetically related to immunoglobulin and TcR domains | + | *** The C- terminus is cytoplasmic |
| − | + | ** Light chain: | |
| − | MHC class I molecules are folded to form specific 3-dimensional structures | + | *** Known as β2-microglobulin |
| + | *** Similar in structure to one of the heavy chain domains | ||
| + | *** Not membrane associated | ||
| + | **** But binds to the α3-domain of the heavy chain | ||
| + | [[Image:MHC I Structure.jpg|thumb|right|150px| Structure of MHC I molecule - Copyright Prof Dirk Werling DrMedVet PhD MRCVS]] | ||
| + | * The MHC class I domains are structurally and genetically related to immunoglobulin and TcR domains | ||
| + | ** The outer domains (α1 and α2) are like the variable domains | ||
| + | ** The α3 domain and β2m are like thrconstant domains | ||
| + | * MHC class I molecules are folded to form specific 3-dimensional structures | ||
| + | ** The α1 and α2 domains are folded to produce an antigen-binding groove | ||
| + | *** This groove can bind molecules of a limited size only (8-10 amino acids) | ||
| + | **** This limits the size of epitope seen by the T-cell receptors | ||
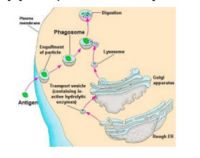
===Presentation Pathway=== | ===Presentation Pathway=== | ||
| − | + | *MHC I presents '''endogenous''' (that is, intracellular) peptides | |
| − | MHC I presents '''endogenous''' (intracellular) peptides | + | *Viral proteins are broken down to peptides by the proteasome and transferred to the endoplasmic reticulum via TAP (Transporters associated with Antigen Processing) molecules |
| + | *In the ER peptides are processed with empty MHC I molecules and exported to the cell surface for presentation | ||
| + | * MHC class I molecules present these to the T-cell receptors of [[Lymphocytes#Cytotoxic CD8+|'''CD8+ T-cells''']] | ||
==MHC II== | ==MHC II== | ||
| − | + | [[Image:MHC II presentation.jpg|thumb|200px|right|'''MHC II presentation, courtesy of Janeway, et al. 2008''']] | |
===Structure=== | ===Structure=== | ||
| − | MHC class II is expressed mainly on [[Macrophages|macrophages]], [[T cell differentiation#Antigen Presentation by Dendritic Cells|dendritic cells]] and [[B | + | * MHC class II is expressed mainly on [[Macrophages|macrophages]], [[T cell differentiation - WikiBlood#Antigen Presentation by Dendritic Cells|dendritic cells]] and [[Lymphocytes#B Cells|B-lymphocytes]] |
| − | + | * MHC class II consists of membrane-associated α and β chains | |
| − | The 3-dimensional structure of MHC class II is similar to MHC class I | + | ** Each chain is a transmembrane glycoprotein |
| − | + | ** The extracellular parts of each chain have two Ig-like domains | |
| + | *** α1 and 7alpha;2, β1 and β2 | ||
| + | **** The outer domains (α1 and β1) are variable-like | ||
| + | **** The inner domains (α2 and β2) are constant-like | ||
| + | * The 3-dimensional structure of MHC class II is similar to MHC class I | ||
| + | ** The outer domains of the α and β chains fold in a similar way to the α1 and α2 domains of class I | ||
| + | *** Produce the antigen-binding groove | ||
| + | [[Image:MHC II Structure.jpg|thumb|right|150px| Structure of MHC II molecule - Copyright Prof Dirk Werling DrMedVet PhD MRCVS]] | ||
===Presentation Pathway=== | ===Presentation Pathway=== | ||
| − | + | *MHC II presents '''exogenous''' (that is, derived from the ECF) peptides | |
| − | MHC II presents '''exogenous''' ( | + | *Endocytosed antigen interacts with MHC II in the cytoplasm to form a complex: |
| − | *Antigen is endoycotsed from the ECF | + | **Antigen is endoycotsed from the ECF |
| − | *Lysosomes fuse with primary endosomes to digest the antigen to peptides | + | **Lysosomes fuse with primary endosomes to digest the antigen to peptides |
| − | *MHC II is meanwhile being produced by the endoplasmic reticulum, along with an invariant chain chaperone | + | **MHC II is meanwhile being produced by the endoplasmic reticulum, along with an invariant chain chaperone |
| − | *These pathways (endoytotic and secretory) merge to allow interaction between the antigen and MHC II | + | **These pathways (endoytotic and secretory) merge to allow interaction between the antigen and MHC II: |
| − | *Foreign antigen then replaces the CLIP peptide | + | ***The invariant chain is digested, leaving a CLIP peptide in the binding groove |
| − | *The MHC II-antigen complex is then secreted to the cell surface for presentation to [[ | + | ***Foreign antigen then replaces the CLIP peptide |
| − | + | *The MHC II-antigen complex is then secreted to the cell surface for presentation to [[Lymphocytes#Helper CD4+|CD4+ T-cells]] | |
| − | |||
| − | |||
| − | Interactions of individual amino acids at the head and tail of the peptide-binding groove control the binding of peptides | + | =Interaction of MHC With Antigen= |
| − | + | * The MHC molecules do not recognise specific amino acid sequences of antigens | |
| − | MHC molecules have the capacity to bind to trillions of different peptides | + | ** Instead, they recognise particular motifs of amino acids |
| + | * The association of any MHC allele with a peptide may be determined by the presence of as few as two amino acids | ||
| + | ** However, these determinants must be present within a particular array | ||
| + | * The actual identity of the amino acids in not important for MHC binding | ||
| + | ** Instead, the physical and chemical characteristics of the amino acid are vital | ||
| + | * Interactions of individual amino acids at the head and tail of the peptide-binding groove control the binding of peptides | ||
| + | ** Are mainly positioned at the floor of the antigen-binding groove, or within the helices facing into the groove | ||
| + | ** These MHC amino acids associate with amino acids near the ends of the peptides | ||
| + | *** The intervening stretch of peptide folds into a helix within the groove | ||
| + | *** Is the target for [[Lymphocytes#T Cells|T cell]] receptor recognition | ||
| + | *MHC molecules have the capacity to bind to trillions of different peptides | ||
| + | **Adopt a flexible '''floppy''' conformation until a peptide binds | ||
| + | **Folds around the peptide to increase stability of the complex | ||
| + | **Uses a small number of anchor residues to tether the peptide allowing different sequences between anchors and different lengths of peptides to bind | ||
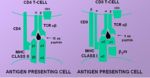
==TCR-MHC Interaction== | ==TCR-MHC Interaction== | ||
| − | [[Image:MHC T cell Interaction.jpg|thumb|right| | + | [[Image:MHC T cell Interaction.jpg|thumb|right|150px|Molecules of T lymphocyte recognition - Copyright Prof Dirk Werling DrMedVet PhD MRCVS]] |
| − | Only peptide associated with MHC will interact with and activate [[T cells]] | + | * Only peptide associated with self-MHC will interact with and activate [[Lymphocytes#T Cells|T cells]] |
| + | ** [[Lymphocytes#T Cells|T cells]] cannot be activated by a peptide on a foreign cell | ||
| + | ** [[Lymphocytes#T Cells|T cells]] will react against foreign MHC molecules | ||
| + | *** This is the basis of graft rejection | ||
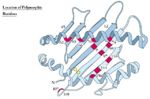
| − | + | [[Image:Location of Polymorphic Residues 1.jpg|thumb|right|150px|Location of Polymorphic Residues - Copyright Prof Dirk Werling DrMedVet PhD MRCVS]] | |
| − | = | + | =The Genetics of the MHC (Polymorphism)= |
| − | + | *Each individual has 6 types of MHC | |
| − | Each individual has 6 types of MHC | + | *MHC molecules are co-dominantly expressed |
| − | + | *The combination of alleles in a chromosome is called an '''MHC Haplotype''' | |
| − | Different individuals have different critical amino acids within the MHC | + | * Different individuals have different critical amino acids within the MHC |
| − | *Each polymorphic variant is called an '''allele''' | + | ** I.e. different amino acids that determine peptide binding |
| − | Both type I and type II MHC molecules are highly polymorphic | + | ** This variation is termed '''MHC polymorphism''' |
| − | + | **Each polymorphic variant is called an '''allele''' | |
| − | Most polymorphisms are point mutations | + | *Both type I and type II MHC molecules are highly polymorphic |
| + | **Most polymorphic regions of class I are in the alpha 1 and alpha 2 domains | ||
| + | **Most polymorphic regions of class II are in the alpha 1 and beta 1 domains | ||
| + | *Most polymorphisms are point mutations | ||
| + | * There are millions of variations in [[Immunoglobulins|antibodies]] and TCR | ||
| + | ** However, with MHC there is very limited variation between molecules | ||
| + | *Allelic variation within the MHC molecule occurs at the peptide binding site and on the top or sides of the binding cleft | ||
| + | *Polymorphisms and polygenism in the MHC protects the population from pathogens evading the immune system | ||
* MHC polymorphism has been best studied in the human | * MHC polymorphism has been best studied in the human | ||
| − | [[Image:Location of Polymorphic Residues 2.jpg|thumb|right| | + | [[Image:Location of Polymorphic Residues 2.jpg|thumb|right|150px|Location of Polymorphic Residues - Copyright Prof Dirk Werling DrMedVet PhD MRCVS]] |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | ==In the Human== | ||
| + | * Humans express: | ||
| + | ** Three types (loci) of MHC class I molecules | ||
| + | *** HLA (Human Leukocyte Antigen)- A, B, and C | ||
| + | ** Three loci of MHC class II molecules | ||
| + | *** HLA-DP, DQ and DR | ||
| + | * In the entire human population there are only approximately 50 different variants (alleles) at each MHC class I and class II locus | ||
| + | ** The variation within MHC class I is entirely on the class I heavy chain | ||
| + | *** The β2m is invariant | ||
| + | ** The variation within MHC class II is mainly within the β chains | ||
| + | * Every individual has two alleles at each MHC locus | ||
| + | ** One inherited from each parent | ||
| + | ** Any individual will therfore express two variants at most at each locus | ||
| + | *** This gives a maximum variability for an individual of: | ||
| + | **** 6 different variants of MHC class I | ||
| + | ***** 2 each of HLA- A, B and C | ||
| + | **** 6 different variants of MHC class II | ||
| + | ***** 2 each of HLA- DP, DQ and DR | ||
| + | * Many animal species have fewer loci than the human | ||
| + | ** E.g. ruminants have no MHC class II DP | ||
| − | + | =MHC and Disease= | |
| − | + | * Antigen from a pathogen has to be seen by the host MHC before an efficient immune response can occur | |
| − | + | ** There is therefore a constant evolutionary battle between the host and the pathogen | |
| − | + | *** There is selective pressure on the pathogen to evolve proteins that do not interact with the host MHC | |
| + | *** There is selective pressure on the host to continue to recognize the pathogen | ||
| + | * The consequence of this parallel evolution is that host-pathogen relationships can lead to the selection of particular MHC variants, for example: | ||
| + | ** MHC class II alleles DR13/DR1*1301 are prevalent in Central and Western Africa | ||
| + | *** Impart resistance to malaria | ||
| + | ** MHC-DRB1 is prevalent in Western Europe, but rare in the Inuit populations of North America | ||
| + | *** Associated with the clearance of hepatitis B infection in Western Europe | ||
| + | *** Inuits have the highest incidence of hepatitis B in the world | ||
| + | ** In humans there are also strong associations between certain alleles and some autoimmune diseases, for example: | ||
| + | *** Diabetes mellitus | ||
| + | *** Ankylosing spondylitis | ||
| + | *** Rheumatoid arthritis | ||
Revision as of 12:51, 13 August 2010
|
|
Introduction
T-cells rely on Major Histocompatability Complexes (MHC) to present antigen fragments for their recognition. MHC has evolved to form two classes for antigen presentation: MHC I presents digestion fragments from antigen in cellular cytoplasm, and MHC II presents digestion fragments from antigen in the tissue fluid. As such, MHC I tends to bind slightly smaller peptides (~9 amino acids) than MHC II (~15 amino acids).
Classes
MHC I
Structure
- MHC class I is expressed on virtually all nucleated cells
- MHC class I consists of a membrane-associated heavy chain bound non-covalently with a secreted light chain
- Heavy chain:
- Made up of three distinct extracellular protein domains
- α1, α2 and α3
- The C- terminus is cytoplasmic
- Made up of three distinct extracellular protein domains
- Light chain:
- Known as β2-microglobulin
- Similar in structure to one of the heavy chain domains
- Not membrane associated
- But binds to the α3-domain of the heavy chain
- Heavy chain:
- The MHC class I domains are structurally and genetically related to immunoglobulin and TcR domains
- The outer domains (α1 and α2) are like the variable domains
- The α3 domain and β2m are like thrconstant domains
- MHC class I molecules are folded to form specific 3-dimensional structures
- The α1 and α2 domains are folded to produce an antigen-binding groove
- This groove can bind molecules of a limited size only (8-10 amino acids)
- This limits the size of epitope seen by the T-cell receptors
- This groove can bind molecules of a limited size only (8-10 amino acids)
- The α1 and α2 domains are folded to produce an antigen-binding groove
Presentation Pathway
- MHC I presents endogenous (that is, intracellular) peptides
- Viral proteins are broken down to peptides by the proteasome and transferred to the endoplasmic reticulum via TAP (Transporters associated with Antigen Processing) molecules
- In the ER peptides are processed with empty MHC I molecules and exported to the cell surface for presentation
- MHC class I molecules present these to the T-cell receptors of CD8+ T-cells
MHC II
Structure
- MHC class II is expressed mainly on macrophages, dendritic cells and B-lymphocytes
- MHC class II consists of membrane-associated α and β chains
- Each chain is a transmembrane glycoprotein
- The extracellular parts of each chain have two Ig-like domains
- α1 and 7alpha;2, β1 and β2
- The outer domains (α1 and β1) are variable-like
- The inner domains (α2 and β2) are constant-like
- α1 and 7alpha;2, β1 and β2
- The 3-dimensional structure of MHC class II is similar to MHC class I
- The outer domains of the α and β chains fold in a similar way to the α1 and α2 domains of class I
- Produce the antigen-binding groove
- The outer domains of the α and β chains fold in a similar way to the α1 and α2 domains of class I
Presentation Pathway
- MHC II presents exogenous (that is, derived from the ECF) peptides
- Endocytosed antigen interacts with MHC II in the cytoplasm to form a complex:
- Antigen is endoycotsed from the ECF
- Lysosomes fuse with primary endosomes to digest the antigen to peptides
- MHC II is meanwhile being produced by the endoplasmic reticulum, along with an invariant chain chaperone
- These pathways (endoytotic and secretory) merge to allow interaction between the antigen and MHC II:
- The invariant chain is digested, leaving a CLIP peptide in the binding groove
- Foreign antigen then replaces the CLIP peptide
- The MHC II-antigen complex is then secreted to the cell surface for presentation to CD4+ T-cells
Interaction of MHC With Antigen
- The MHC molecules do not recognise specific amino acid sequences of antigens
- Instead, they recognise particular motifs of amino acids
- The association of any MHC allele with a peptide may be determined by the presence of as few as two amino acids
- However, these determinants must be present within a particular array
- The actual identity of the amino acids in not important for MHC binding
- Instead, the physical and chemical characteristics of the amino acid are vital
- Interactions of individual amino acids at the head and tail of the peptide-binding groove control the binding of peptides
- Are mainly positioned at the floor of the antigen-binding groove, or within the helices facing into the groove
- These MHC amino acids associate with amino acids near the ends of the peptides
- The intervening stretch of peptide folds into a helix within the groove
- Is the target for T cell receptor recognition
- MHC molecules have the capacity to bind to trillions of different peptides
- Adopt a flexible floppy conformation until a peptide binds
- Folds around the peptide to increase stability of the complex
- Uses a small number of anchor residues to tether the peptide allowing different sequences between anchors and different lengths of peptides to bind
TCR-MHC Interaction
- Only peptide associated with self-MHC will interact with and activate T cells
The Genetics of the MHC (Polymorphism)
- Each individual has 6 types of MHC
- MHC molecules are co-dominantly expressed
- The combination of alleles in a chromosome is called an MHC Haplotype
- Different individuals have different critical amino acids within the MHC
- I.e. different amino acids that determine peptide binding
- This variation is termed MHC polymorphism
- Each polymorphic variant is called an allele
- Both type I and type II MHC molecules are highly polymorphic
- Most polymorphic regions of class I are in the alpha 1 and alpha 2 domains
- Most polymorphic regions of class II are in the alpha 1 and beta 1 domains
- Most polymorphisms are point mutations
- There are millions of variations in antibodies and TCR
- However, with MHC there is very limited variation between molecules
- Allelic variation within the MHC molecule occurs at the peptide binding site and on the top or sides of the binding cleft
- Polymorphisms and polygenism in the MHC protects the population from pathogens evading the immune system
- MHC polymorphism has been best studied in the human
In the Human
- Humans express:
- Three types (loci) of MHC class I molecules
- HLA (Human Leukocyte Antigen)- A, B, and C
- Three loci of MHC class II molecules
- HLA-DP, DQ and DR
- Three types (loci) of MHC class I molecules
- In the entire human population there are only approximately 50 different variants (alleles) at each MHC class I and class II locus
- The variation within MHC class I is entirely on the class I heavy chain
- The β2m is invariant
- The variation within MHC class II is mainly within the β chains
- The variation within MHC class I is entirely on the class I heavy chain
- Every individual has two alleles at each MHC locus
- One inherited from each parent
- Any individual will therfore express two variants at most at each locus
- This gives a maximum variability for an individual of:
- 6 different variants of MHC class I
- 2 each of HLA- A, B and C
- 6 different variants of MHC class II
- 2 each of HLA- DP, DQ and DR
- 6 different variants of MHC class I
- This gives a maximum variability for an individual of:
- Many animal species have fewer loci than the human
- E.g. ruminants have no MHC class II DP
MHC and Disease
- Antigen from a pathogen has to be seen by the host MHC before an efficient immune response can occur
- There is therefore a constant evolutionary battle between the host and the pathogen
- There is selective pressure on the pathogen to evolve proteins that do not interact with the host MHC
- There is selective pressure on the host to continue to recognize the pathogen
- There is therefore a constant evolutionary battle between the host and the pathogen
- The consequence of this parallel evolution is that host-pathogen relationships can lead to the selection of particular MHC variants, for example:
- MHC class II alleles DR13/DR1*1301 are prevalent in Central and Western Africa
- Impart resistance to malaria
- MHC-DRB1 is prevalent in Western Europe, but rare in the Inuit populations of North America
- Associated with the clearance of hepatitis B infection in Western Europe
- Inuits have the highest incidence of hepatitis B in the world
- In humans there are also strong associations between certain alleles and some autoimmune diseases, for example:
- Diabetes mellitus
- Ankylosing spondylitis
- Rheumatoid arthritis
- MHC class II alleles DR13/DR1*1301 are prevalent in Central and Western Africa