Difference between revisions of "Recognition of Microorganisms"
Jump to navigation
Jump to search
Rjfrancisrvc (talk | contribs) |
Rjfrancisrvc (talk | contribs) |
||
| Line 15: | Line 15: | ||
The engagement of PRRs by PAMPs triggers: | The engagement of PRRs by PAMPs triggers: | ||
* '''[[Phagocytosis|Phagocytosis]]''' | * '''[[Phagocytosis|Phagocytosis]]''' | ||
| − | * The expression of '''cytokines''', which brings about [[:Category:Inflammation|inflammation]] and other immune responses | + | * The expression of '''[[Cytokines|cytokines]]''', which brings about [[:Category:Inflammation|inflammation]] and other immune responses |
<br /> | <br /> | ||
<br /> | <br /> | ||
| + | |||
==Pattern Recognition Receptors== | ==Pattern Recognition Receptors== | ||
<big><center>'''''Examples of Pattern Recognition Receptors'''''</center></big> | <big><center>'''''Examples of Pattern Recognition Receptors'''''</center></big> | ||
Revision as of 11:32, 10 May 2012
The innate immune system recognises components of pathogens which are intrinsically foreign (i.e. not present on normal mammalian cells), such as:
- Lipopolysaccharides of gram-negative bacteria
- Peptidoglycans of gram-positive bacteria
- Mannose sugars
- D-isoform amino acids
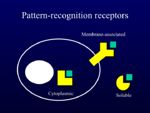
These are known to the body as foreign as they are expressed as pathogen-associated molecular patterns (PAMPs) which are recognised by pattern recognition receptors (PRRs) expressed on mammalian cells. These receptors are expressed on many different cell types, not just on phagocytes, though not all are expressed by all cells: different cell types express a different range of PRRs. PRRs are either intracellular, membrane-associated or soluble:
- Recognition of pathogens via the cellular PRRs results in phagocytosis and inflammation
- Recognition of pathogens via the humoral PRRs results in various killing mechanisms
Actions
The engagement of PRRs by PAMPs triggers:
- Phagocytosis
- The expression of cytokines, which brings about inflammation and other immune responses
Pattern Recognition Receptors
| Receptor | Location | Ligands |
|---|---|---|
| TLR2 (Toll-like receptor) | Cell Membrane | Peptidoglycan of gram +ve bacteria |
| TLR3 | Cell Membrane | dsRNA of RNA viruses (e.g. avian influenza) |
| TLR4 | Cell Membrane | Lipoplysaccharide from gram-negative bacteria (e.g. E. coli, Salmonella) |
| TLR5 | Cell Membrane | Bacterial flagellin |
| TLR9 | Cell Membrane | Bacterial DNA (CpG DNA) |
| C-type lectins | Soluble | Carbohydrates, all bacteria, dead cells |
| fMLP | Soluble | Formyl peptides (i.e. all bacteria) |
| Complement receptors | Soluble | Fixed complement components (e.g. iC3b) |
| NOD2 | Cytoplasm | Peptidoglycan of gram +ve bacteria |
| dsRNA-dependent Protein Kinase Receptor | Cytoplasm | ds RNA of RNA viruses |
| Originally funded by the RVC Jim Bee Award 2007 |