Difference between revisions of "Complement"
Rjfrancisrvc (talk | contribs) |
|||
| (40 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | {{review}} | ||
==Introduction== | ==Introduction== | ||
| − | Complement is so called because it complements the function of | + | Complement is so called because it complements the function of antibody. It is a [[Triggered Enzyme Cascade - WikiBlood|triggered enzyme cascade]] and there are more than 20 different proteins in the complement cascades, with most being enzymes or pro-enzymes. It can be activated by both the innate and adaptive immune systems and is one of the main innate protective mechanisms of invertebrates. Due to its destructive potential the complement system is heavily regulated but when activated it works largely by forming pore complexes as well as triggering acute inflammation and by promoting [[Phagocytosis|phagocytosis]] by [[Macrophages|macrophages]] and [[Neutrophils|neutrophils]]. |
| − | |||
==Complement Fixation Pathways== | ==Complement Fixation Pathways== | ||
| − | [[Image:LH Complement pathway.png|thumb| | + | [[Image:LH Complement pathway.png|thumb|right|250px|'''Classical and alternative pathways''']] |
===Classical Pathway=== | ===Classical Pathway=== | ||
| − | [[Image:Complement Classical Pathway.png|thumb|right|250px|'''Classical pathway in detail''' | + | [[Image:Complement Classical Pathway.png|thumb|right|250px|'''Classical pathway in detail''']] |
| − | + | <p>Triggered by the binding of antibody to antigen. Only [[IgM]] and certain [[IgG]] subclasses can activate complement this way. | |
| − | [[IgM]] and certain [[IgG]]. | + | #Immune complexes trigger complement activation by binding C1. |
| − | + | #*C1 cross-links two antibody molecules | |
| − | + | #*C1 is a complex of: | |
| − | + | #**C1q | |
| − | + | #***Looks like a bunch of 6 tulips | |
| − | + | #***Each "flower" consists of a globular protein head and a collagen "stem" | |
| − | The | + | #***At least two C1q globular heads must bind to antibody before the complement cascade is triggered |
| − | + | #**C1r | |
| − | + | #**C1s | |
| − | + | #*C1r and C1s become activated when at least 2 C1q heads are antibody-bound (to FC region) | |
| − | < | + | #**Form the enzyme C1 esterase (serine protease) |
| − | < | + | #C1 esterase first digests C4 to C4a and C4b and the C2 to C2a and C2b |
| + | #*C4b binds to the antigen | ||
| + | #C4b then binds C2a | ||
| + | #*C4b and C2a complex to form the enzyme C2a¯4b (C3 convertase) | ||
| + | #C2a¯4b digests C3 into C3a and C3b. | ||
| + | #*The production of C3b can now be amplified by the same mechanism as the alternative pathway | ||
| + | #*The binding of one C1q molecule produces one C1 esterase molecule that then causes the binding of many hundreds of C4b¯2b molecules | ||
| + | #The C3b molecule that is produced by the action of this enzyme can also bind to it | ||
| + | #*Forms C4b2¯a3b (C5 convertase) | ||
| + | #*This activates C5 and initiates the [[#Membrane Attack Complex|membrane attack complex]] (MAC) | ||
| + | *Ca fragments C3a, C4a and C5a, and Cb fragment C2b | ||
| + | **Are chemotactic for [[Neutrophils|neutrophils]] | ||
| + | **Induce acute inflammation | ||
| + | **Increase vascular permeability and cell adhesion molecules | ||
| + | ***Cause [[Mast Cells|mast cell]] and [[Basophils|basophil]] degranulation</p> | ||
| + | <p>Like the alternative pathway, another major effect of classical pathway activation is to produce iC3b due to the actions of [[#Factors I and H|Factors I and H]] which promotes phagocytosis and initiates inflammation</p> | ||
===Alternative Pathway=== | ===Alternative Pathway=== | ||
| − | [[Image:Complement Alternative Pathway.png|thumb|right|250px|'''Alternative pathway in detail''' | + | [[Image:Complement Alternative Pathway.png|thumb|right|250px|'''Alternative pathway in detail''']] |
| − | <p> | + | <p>Is found in animals that do not have antibodies i.e. animals lower than cartilaginous fish. It was discovered after the classical pathway and is thus called the 'alternative pathway'. |
| − | + | *C3b has a binding site that anchors it to any surface | |
| − | + | **The surfaces of certain activators stabilise the otherwise short-lived C3b in the absence of antibody | |
| − | + | ***Gram-negative bacteria, yeasts and fungi are the most efficient activators | |
| − | + | *When active C3b is bound to particle surfaces, it is protected from inactivation by another complement component, properdin | |
| − | + | *C3b can then bind Factor B producing the complex C3bB | |
| − | + | *C3bB is the only substrate for a plasma enzyme known as Factor D. | |
| − | + | *Factor D splits a small peptide (the Ba peptide) from Factor B | |
| − | < | + | **This generates C3b¯Bb. (C3 convertase) |
| − | < | + | **C3b¯Bb acts on C3 to generate more C3b |
| − | At the same time as | + | **C3b then generates more C3b¯Bb |
| − | + | ***An efficient positive feedback loop | |
| − | [[Image:Complement Mannose Binding Lectin Pathway.png|thumb|right|250px|'''Mannose Binding Lectin pathway in detail''' | + | *C3b¯Bb can also bind C3b to form C3bB¯b3b. |
| − | < | + | **C3bB¯b3b is one of the two enzymes that activates the [[#Membrane Attack Complex|membrane attack complex]] (MAC) |
| + | *C3bB¯b3b splits C5 into: | ||
| + | **C5a, a small pro-inflammatory peptide | ||
| + | **C5b, the initiator of the MAC</p> | ||
| + | <p> | ||
| + | *At the same time as C3bB¯b3b is being formed, Factors I and H are acting to breakdown C3b to iC3b | ||
| + | **The C3b being broken down may be: | ||
| + | ***As single C3b or in the complex C3b¯Bb. | ||
| + | ***In plasma or on bacterial surfaces. | ||
| + | *iC3b is inactive in the complement cascade but is a major target for phagocytes | ||
| + | **Phagocytes have large numbers of iC3b receptors (complement receptors) which give opsonisation when engaged. </p> | ||
| + | [[Image:Complement Mannose Binding Lectin Pathway.png|thumb|right|250px|'''Mannose Binding Lectin pathway in detail''']] | ||
| + | <p>The main effects of alternative complement activation are therefore: | ||
| + | #To coat bacteria with iC3b | ||
| + | #*A major target for [[Phagocytosis|phagocytosis]] by [[Macrophages|macrophages]] and [[Neutrophils|neutrophils]] via the complement receptors | ||
| + | #To induce an acute inflammatory response via C3a and C5a. | ||
| + | #*Chemotactic for [[Neutrophils|neutrophils]] | ||
| + | #*Induce the production of the cytokines (IL-1β and TNFα) responsible for acute inflammation</p> | ||
===Mannose Binding-Lectin Pathway=== | ===Mannose Binding-Lectin Pathway=== | ||
| − | Lectins are proteins that bind carbohydrates e.g. collectins. They bind | + | <p>Lectins are proteins that bind carbohydrates e.g. collectins. They bind carbohydrates that have a terminal mannose residue and are called mannose-binding lectins. |
| − | + | *Secreted by the liver into plasma. | |
| − | + | * The action of lectin binding to carbohydrate activates plasma-associated proteases called mannose-binding lectin associated proteases (MASPs) | |
| − | The | + | ** These act on C4 and C2 in the same way as C1 esterase, and on C3 in much the same manner as C3 convertase</p> |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | < | ||
==Membrane Attack Complex== | ==Membrane Attack Complex== | ||
[[Image:LH Complement lysis.png|thumb|200px|right|'''Membrane Attack Complex''']] | [[Image:LH Complement lysis.png|thumb|200px|right|'''Membrane Attack Complex''']] | ||
| − | This is the lytic pathway of complement function and can be initiated via either | + | This is the lytic pathway of complement function and can be initiated via either C3bB¯b3b or C4b2¯a3b. |
| − | + | #Split C5 into: | |
| − | + | #*C5a is pro-inflammatory. | |
| − | + | #*C5b the first molecule of the MAC (very short-lived and biologically active) | |
| − | C5b rapidly attaches to cell surfaces and binds to one C6 molecule | + | #C5b rapidly attaches to cell surfaces and binds to one C6 molecule |
| − | + | #C5bC6 binds one C7 molecule | |
| − | < | + | #*C5bC6 binds to cell membrane via C7 |
| + | #C5b67 binds one C8 molecule | ||
| + | #*C8 inserts into the cell membrane | ||
| + | # C9 molecules bind and polymerize within the cell membrane (1-16 C9 molecules)</p> | ||
| + | <p>The polymerization of C9 results in a small pore being formed which causes cell lysis by osmotic shock.</p> | ||
==Complement Inhibitors== | ==Complement Inhibitors== | ||
| − | To prevent inappropriate activation of the complement system there is a range of control mechanisms/ | + | <p>To prevent inappropriate activation of the complement system there is a range of control mechanisms/compunds.</p> |
| − | < | ||
=====Decay accelerating factor (DAF)===== | =====Decay accelerating factor (DAF)===== | ||
Is both secreted onto and present on cell membranes and hastens the degradation of C1 esterase. Exerts control on the [[#Classical Pathway|classical pathway]]. | Is both secreted onto and present on cell membranes and hastens the degradation of C1 esterase. Exerts control on the [[#Classical Pathway|classical pathway]]. | ||
| Line 73: | Line 100: | ||
=====CD59===== | =====CD59===== | ||
This binds the first molecule of C9 when it inserts into a cell membrane and prevents the polymerisation of C9 and therefore pore formation and cell lysis. It acts as a protective mechanism for the host cells. | This binds the first molecule of C9 when it inserts into a cell membrane and prevents the polymerisation of C9 and therefore pore formation and cell lysis. It acts as a protective mechanism for the host cells. | ||
| − | |||
==Functions== | ==Functions== | ||
===Opsonisation=== | ===Opsonisation=== | ||
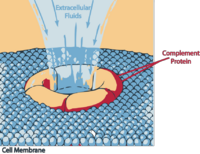
| − | Once activated the complement system deposits a shell of protein on the bacterial cell surface. C4b, C3b, C5b and C7 molecules contain active binding sites that anchor the complex to the surface and the major protein on the cell surface is iC3b | + | <p>Once activated the complement system deposits a shell of protein on the bacterial cell surface. C4b, C3b, C5b and C7 molecules contain active binding sites that anchor the complex to the surface and the major protein on the cell surface is iC3b (some of the smaller C3 breakdown products (e.g. C3d) are also present).</p> |
| − | + | <p>Complement fragments (C2b, C3a, C4a and especially C5a) released after complement activation are chemotactic for phagocytes and iC3b acts as a target for [[Phagocytosis|phagocytosis]]. Phagocytes have receptors that bind avidly with the complement fragments. Complement-mediated opsonisation of microorganisms is several thousand times more efficient that innate receptors.</p> | |
| − | Complement fragments (C2b, C3a, C4a and especially C5a) released after complement activation are chemotactic for phagocytes and iC3b acts as a target for [[Phagocytosis|phagocytosis]]. Phagocytes have receptors that bind avidly with the complement fragments. Complement-mediated opsonisation of microorganisms is several thousand times more efficient | ||
===Inflammation=== | ===Inflammation=== | ||
| − | The smaller complement peptides are very efficient at inducing inflammation i.e. C3a and C5a. They attract [[Blood Cells - Overview|granulocytes]] to the site of complement activation as well as stimulating their degranulation.</p> | + | <p>The smaller complement peptides are very efficient at inducing inflammation i.e. C3a and C5a. They attract [[Blood Cells - Overview|granulocytes]] to the site of complement activation as well as stimulating their degranulation.</p> |
===Cell Lysis=== | ===Cell Lysis=== | ||
<p>The components (C5 – C9) can kill pathogens directly by causing cell lysis. This is effective against encapsulated bacterial infection like Neisseria and Meningococci.</p> | <p>The components (C5 – C9) can kill pathogens directly by causing cell lysis. This is effective against encapsulated bacterial infection like Neisseria and Meningococci.</p> | ||
| − | |||
==Links== | ==Links== | ||
*[[Complement Associated Diseases|Pathology of complement associated diseases]] | *[[Complement Associated Diseases|Pathology of complement associated diseases]] | ||
| Line 91: | Line 115: | ||
{{Learning | {{Learning | ||
|flashcards = [[Complement Flashcards]] | |flashcards = [[Complement Flashcards]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
}} | }} | ||
<br><br> | <br><br> | ||
| − | |||
| − | |||
| − | |||
{{Jim Bee 2007}} | {{Jim Bee 2007}} | ||
[[Category:Innate Immune System|C]] | [[Category:Innate Immune System|C]] | ||
[[Category:Image Review]] | [[Category:Image Review]] | ||
| − | |||
Revision as of 10:13, 27 April 2012
| This article has been peer reviewed but is awaiting expert review. If you would like to help with this, please see more information about expert reviewing. |
Introduction
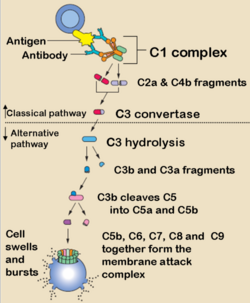
Complement is so called because it complements the function of antibody. It is a triggered enzyme cascade and there are more than 20 different proteins in the complement cascades, with most being enzymes or pro-enzymes. It can be activated by both the innate and adaptive immune systems and is one of the main innate protective mechanisms of invertebrates. Due to its destructive potential the complement system is heavily regulated but when activated it works largely by forming pore complexes as well as triggering acute inflammation and by promoting phagocytosis by macrophages and neutrophils.
Complement Fixation Pathways
Classical Pathway
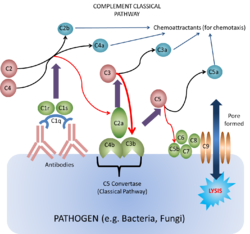
Triggered by the binding of antibody to antigen. Only IgM and certain IgG subclasses can activate complement this way.
- Immune complexes trigger complement activation by binding C1.
- C1 cross-links two antibody molecules
- C1 is a complex of:
- C1q
- Looks like a bunch of 6 tulips
- Each "flower" consists of a globular protein head and a collagen "stem"
- At least two C1q globular heads must bind to antibody before the complement cascade is triggered
- C1r
- C1s
- C1q
- C1r and C1s become activated when at least 2 C1q heads are antibody-bound (to FC region)
- Form the enzyme C1 esterase (serine protease)
- C1 esterase first digests C4 to C4a and C4b and the C2 to C2a and C2b
- C4b binds to the antigen
- C4b then binds C2a
- C4b and C2a complex to form the enzyme C2a¯4b (C3 convertase)
- C2a¯4b digests C3 into C3a and C3b.
- The production of C3b can now be amplified by the same mechanism as the alternative pathway
- The binding of one C1q molecule produces one C1 esterase molecule that then causes the binding of many hundreds of C4b¯2b molecules
- The C3b molecule that is produced by the action of this enzyme can also bind to it
- Forms C4b2¯a3b (C5 convertase)
- This activates C5 and initiates the membrane attack complex (MAC)
- Ca fragments C3a, C4a and C5a, and Cb fragment C2b
- Are chemotactic for neutrophils
- Induce acute inflammation
- Increase vascular permeability and cell adhesion molecules
Like the alternative pathway, another major effect of classical pathway activation is to produce iC3b due to the actions of Factors I and H which promotes phagocytosis and initiates inflammation
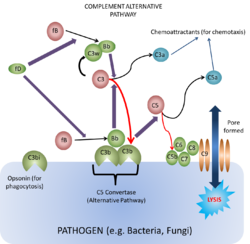
Alternative Pathway
Is found in animals that do not have antibodies i.e. animals lower than cartilaginous fish. It was discovered after the classical pathway and is thus called the 'alternative pathway'.
- C3b has a binding site that anchors it to any surface
- The surfaces of certain activators stabilise the otherwise short-lived C3b in the absence of antibody
- Gram-negative bacteria, yeasts and fungi are the most efficient activators
- The surfaces of certain activators stabilise the otherwise short-lived C3b in the absence of antibody
- When active C3b is bound to particle surfaces, it is protected from inactivation by another complement component, properdin
- C3b can then bind Factor B producing the complex C3bB
- C3bB is the only substrate for a plasma enzyme known as Factor D.
- Factor D splits a small peptide (the Ba peptide) from Factor B
- This generates C3b¯Bb. (C3 convertase)
- C3b¯Bb acts on C3 to generate more C3b
- C3b then generates more C3b¯Bb
- An efficient positive feedback loop
- C3b¯Bb can also bind C3b to form C3bB¯b3b.
- C3bB¯b3b is one of the two enzymes that activates the membrane attack complex (MAC)
- C3bB¯b3b splits C5 into:
- C5a, a small pro-inflammatory peptide
- C5b, the initiator of the MAC
- At the same time as C3bB¯b3b is being formed, Factors I and H are acting to breakdown C3b to iC3b
- The C3b being broken down may be:
- As single C3b or in the complex C3b¯Bb.
- In plasma or on bacterial surfaces.
- The C3b being broken down may be:
- iC3b is inactive in the complement cascade but is a major target for phagocytes
- Phagocytes have large numbers of iC3b receptors (complement receptors) which give opsonisation when engaged.
The main effects of alternative complement activation are therefore:
- To coat bacteria with iC3b
- A major target for phagocytosis by macrophages and neutrophils via the complement receptors
- To induce an acute inflammatory response via C3a and C5a.
- Chemotactic for neutrophils
- Induce the production of the cytokines (IL-1β and TNFα) responsible for acute inflammation
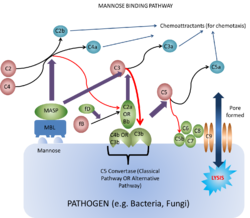
Mannose Binding-Lectin Pathway
Lectins are proteins that bind carbohydrates e.g. collectins. They bind carbohydrates that have a terminal mannose residue and are called mannose-binding lectins.
- Secreted by the liver into plasma.
- The action of lectin binding to carbohydrate activates plasma-associated proteases called mannose-binding lectin associated proteases (MASPs)
- These act on C4 and C2 in the same way as C1 esterase, and on C3 in much the same manner as C3 convertase
Membrane Attack Complex
This is the lytic pathway of complement function and can be initiated via either C3bB¯b3b or C4b2¯a3b.
- Split C5 into:
- C5a is pro-inflammatory.
- C5b the first molecule of the MAC (very short-lived and biologically active)
- C5b rapidly attaches to cell surfaces and binds to one C6 molecule
- C5bC6 binds one C7 molecule
- C5bC6 binds to cell membrane via C7
- C5b67 binds one C8 molecule
- C8 inserts into the cell membrane
- C9 molecules bind and polymerize within the cell membrane (1-16 C9 molecules)
The polymerization of C9 results in a small pore being formed which causes cell lysis by osmotic shock.
Complement Inhibitors
To prevent inappropriate activation of the complement system there is a range of control mechanisms/compunds.
Decay accelerating factor (DAF)
Is both secreted onto and present on cell membranes and hastens the degradation of C1 esterase. Exerts control on the classical pathway.
Factors I and H
Break down C3b and controls positive feedback by inhibiting C3b¯Bb thus preventing the complement cascade from running to exhaustion each time it is activated.
Complement receptor 1 (CR1)
Is present on many cell types especially erythrocytes and functions to bind C3d (the breakdown product of C3b) resulting from the action of Factors I and H. It binds potentially inflammatory immune complexes in plasma and these are then transported to the liver where they are phagocytosed by the hepatic macrophages and removed.
CD59
This binds the first molecule of C9 when it inserts into a cell membrane and prevents the polymerisation of C9 and therefore pore formation and cell lysis. It acts as a protective mechanism for the host cells.
Functions
Opsonisation
Once activated the complement system deposits a shell of protein on the bacterial cell surface. C4b, C3b, C5b and C7 molecules contain active binding sites that anchor the complex to the surface and the major protein on the cell surface is iC3b (some of the smaller C3 breakdown products (e.g. C3d) are also present).
Complement fragments (C2b, C3a, C4a and especially C5a) released after complement activation are chemotactic for phagocytes and iC3b acts as a target for phagocytosis. Phagocytes have receptors that bind avidly with the complement fragments. Complement-mediated opsonisation of microorganisms is several thousand times more efficient that innate receptors.
Inflammation
The smaller complement peptides are very efficient at inducing inflammation i.e. C3a and C5a. They attract granulocytes to the site of complement activation as well as stimulating their degranulation.
Cell Lysis
The components (C5 – C9) can kill pathogens directly by causing cell lysis. This is effective against encapsulated bacterial infection like Neisseria and Meningococci.
Links
| Complement Learning Resources | |
|---|---|
 Test your knowledge using flashcard type questions |
Complement Flashcards |
| Originally funded by the RVC Jim Bee Award 2007 |